Correction of batch/block differences.
Arguments
- d
S4 object of class
AnalysisData- block
sample information column name to use containing sample block groupings
- type
type of average to use
Details
There can sometimes be artificial batch related variability introduced into metabolomics analyses as a result of analytical instrumentation or sample preparation. With an appropriate randomised block design of sample injection order, batch related variability can be corrected using an average centring correction method of the individual features.
Examples
## Initial example data preparation
library(metaboData)
d <- analysisData(abr1$neg[,200:300],abr1$fact) %>%
occupancyMaximum(occupancy = 2/3)
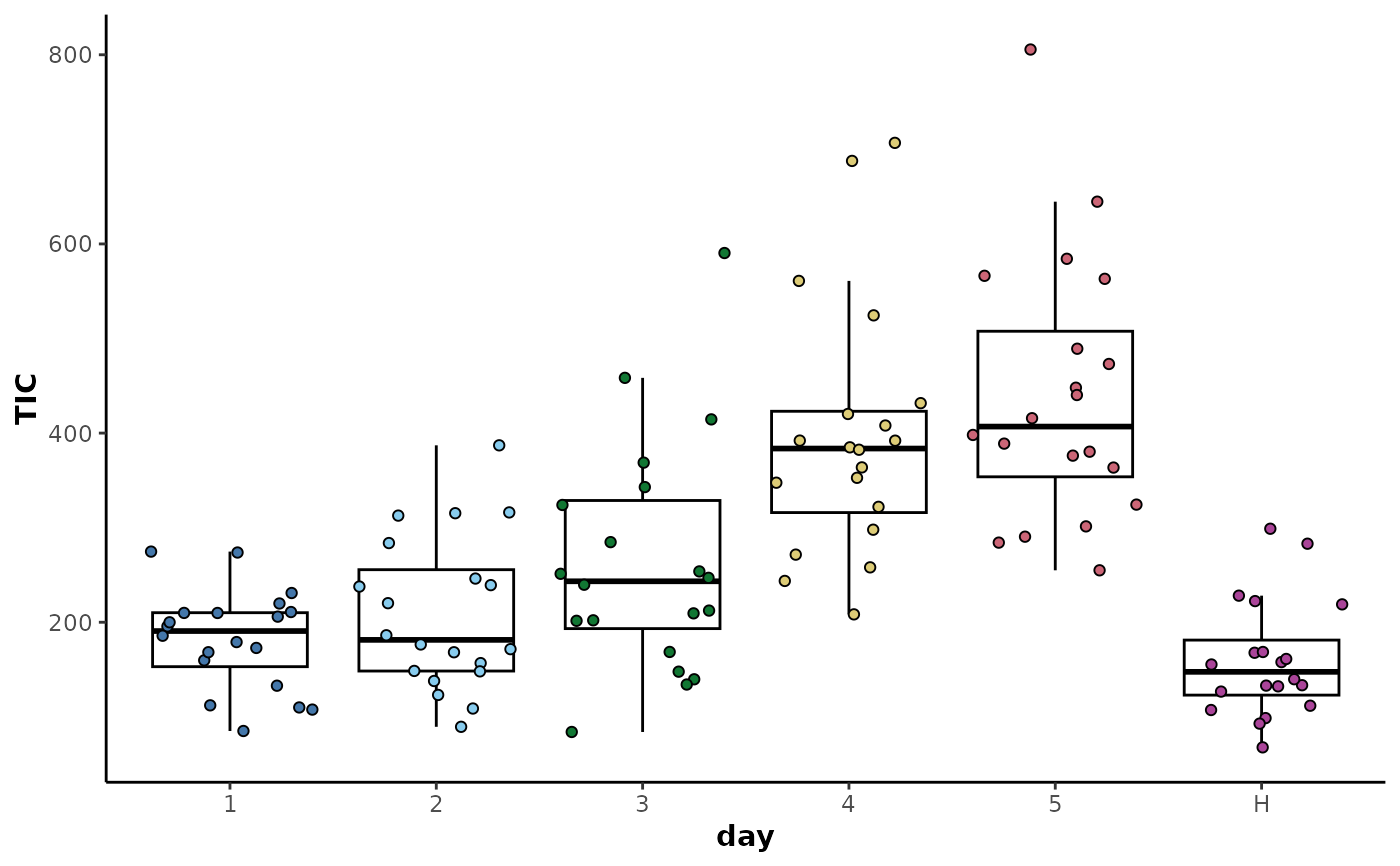
## Group total ion count distributions prior to correction
d %>%
plotTIC(by = 'day',colour = 'day')
 ## Group total ion count distributions after group median correction
d %>%
correctionCenter(block = 'day',type = 'median') %>%
plotTIC(by = 'day',colour = 'day')
## Group total ion count distributions after group median correction
d %>%
correctionCenter(block = 'day',type = 'median') %>%
plotTIC(by = 'day',colour = 'day')
