Impute missing values using random forest imputation.
Usage
imputeAll(d, occupancy = 2/3, parallel = "variables", seed = 1234)
# S4 method for AnalysisData
imputeAll(d, occupancy = 2/3, parallel = "variables", seed = 1234)
imputeClass(d, cls = "class", occupancy = 2/3, seed = 1234)
# S4 method for AnalysisData
imputeClass(d, cls = "class", occupancy = 2/3, seed = 1234)Arguments
- d
S4 object of class
AnalysisData- occupancy
occupancy threshold above which missing values of a feature will be imputed
- parallel
parallel type to use. See
?missForestfor details- seed
random number seed
- cls
info column to use for class labels
Details
Missing values can have an important influence on downstream analyses with zero values heavily influencing the outcomes of parametric tests.
Where and how they are imputed are important considerations and is highly related to variable occupancy.
The methods provided here allow both these aspects to be taken into account and utilise random forest imputation using the missForest package.
Methods
imputeAll: Impute missing values across all sample features.imputeClass: Impute missing values class-wise.
Examples
## Each of the following examples shows the application of each imputation method and then
## a Linear Discriminant Analysis is plotted to show it's effect on the data structure.
## Initial example data preparation
library(metaboData)
d <- analysisData(abr1$neg[,200:250],abr1$fact) %>%
occupancyMaximum(occupancy = 2/3)
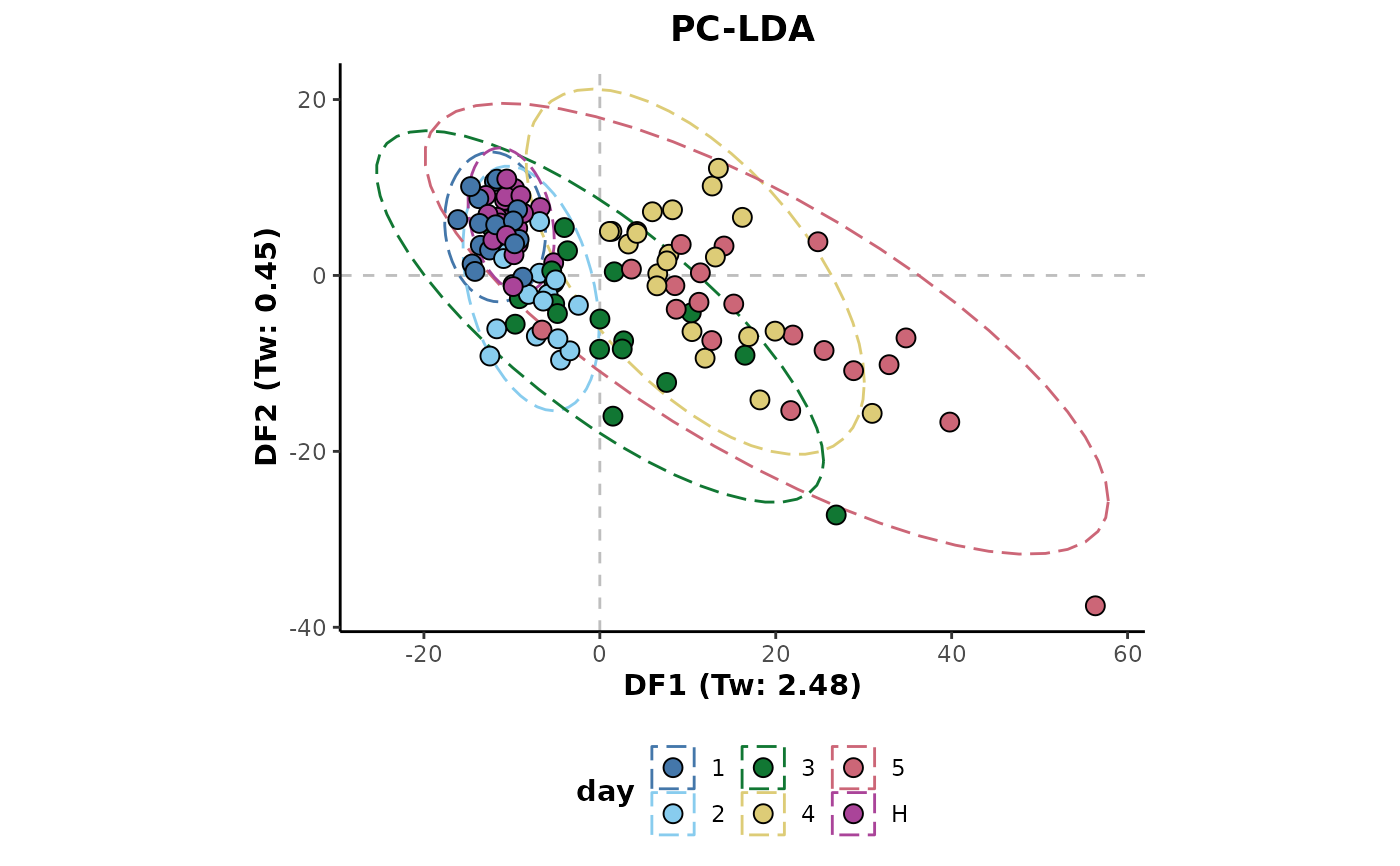
d %>%
plotLDA(cls = 'day')
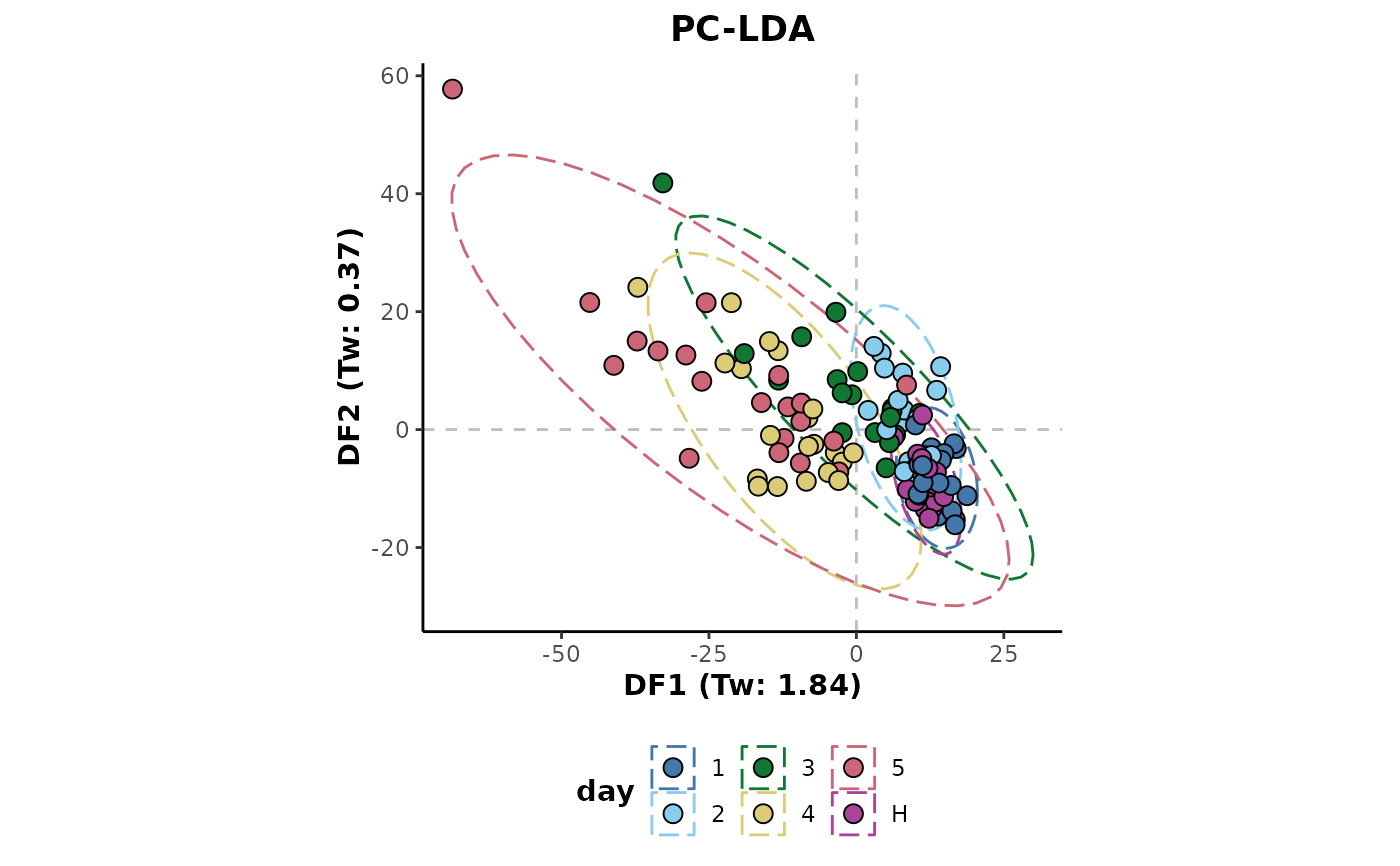 ## Missing value imputation across all samples
d %>%
imputeAll(parallel = 'no') %>%
plotLDA(cls = 'day')
## Missing value imputation across all samples
d %>%
imputeAll(parallel = 'no') %>%
plotLDA(cls = 'day')
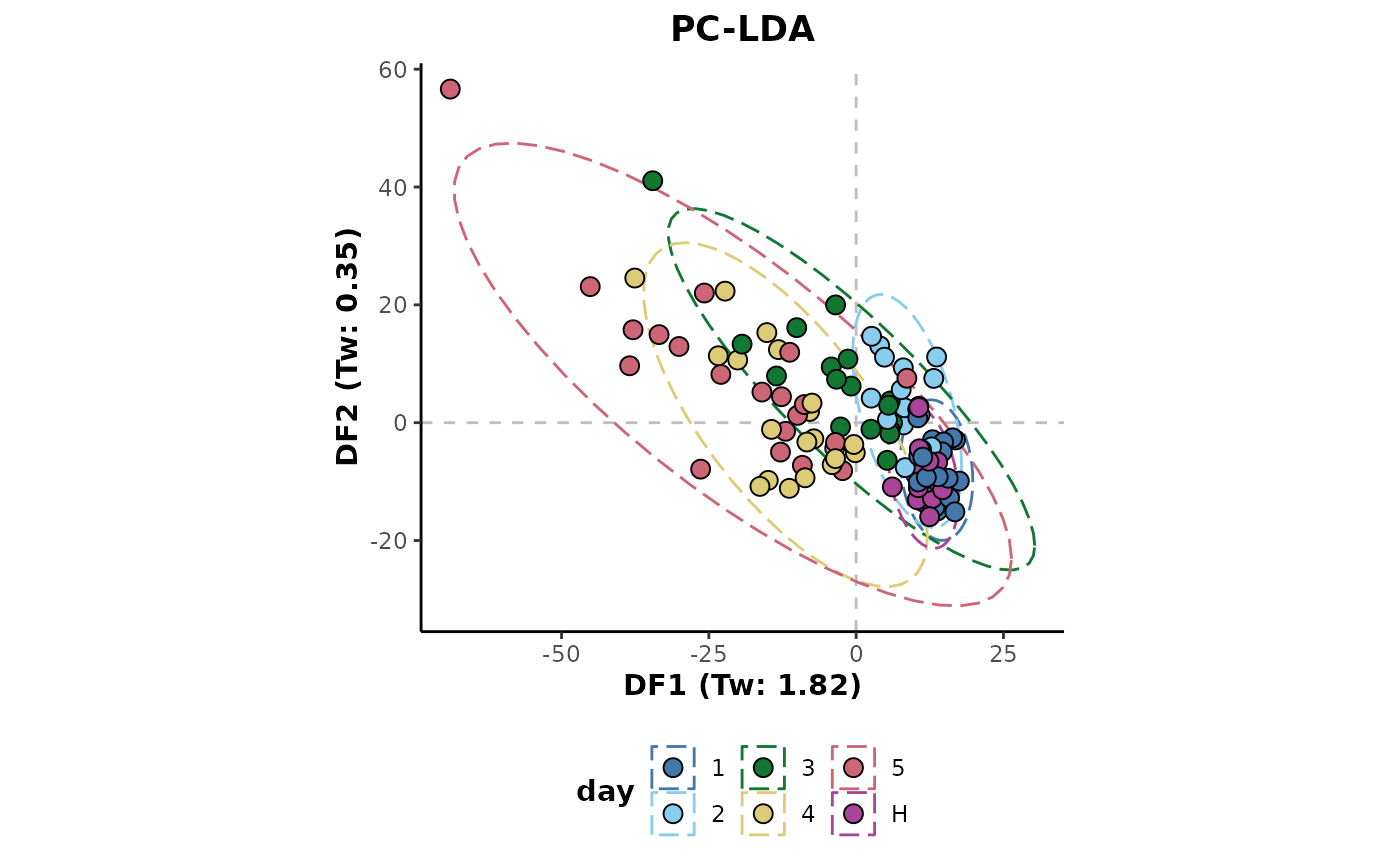 ## Missing value imputation class-wise
d %>%
imputeClass(cls = 'day') %>%
plotLDA(cls = 'day')
## Missing value imputation class-wise
d %>%
imputeClass(cls = 'day') %>%
plotLDA(cls = 'day')